Dr. Marvin J. Bayro
 Protein structure and dynamics in molecular therapeutics
Protein structure and dynamics in molecular therapeutics
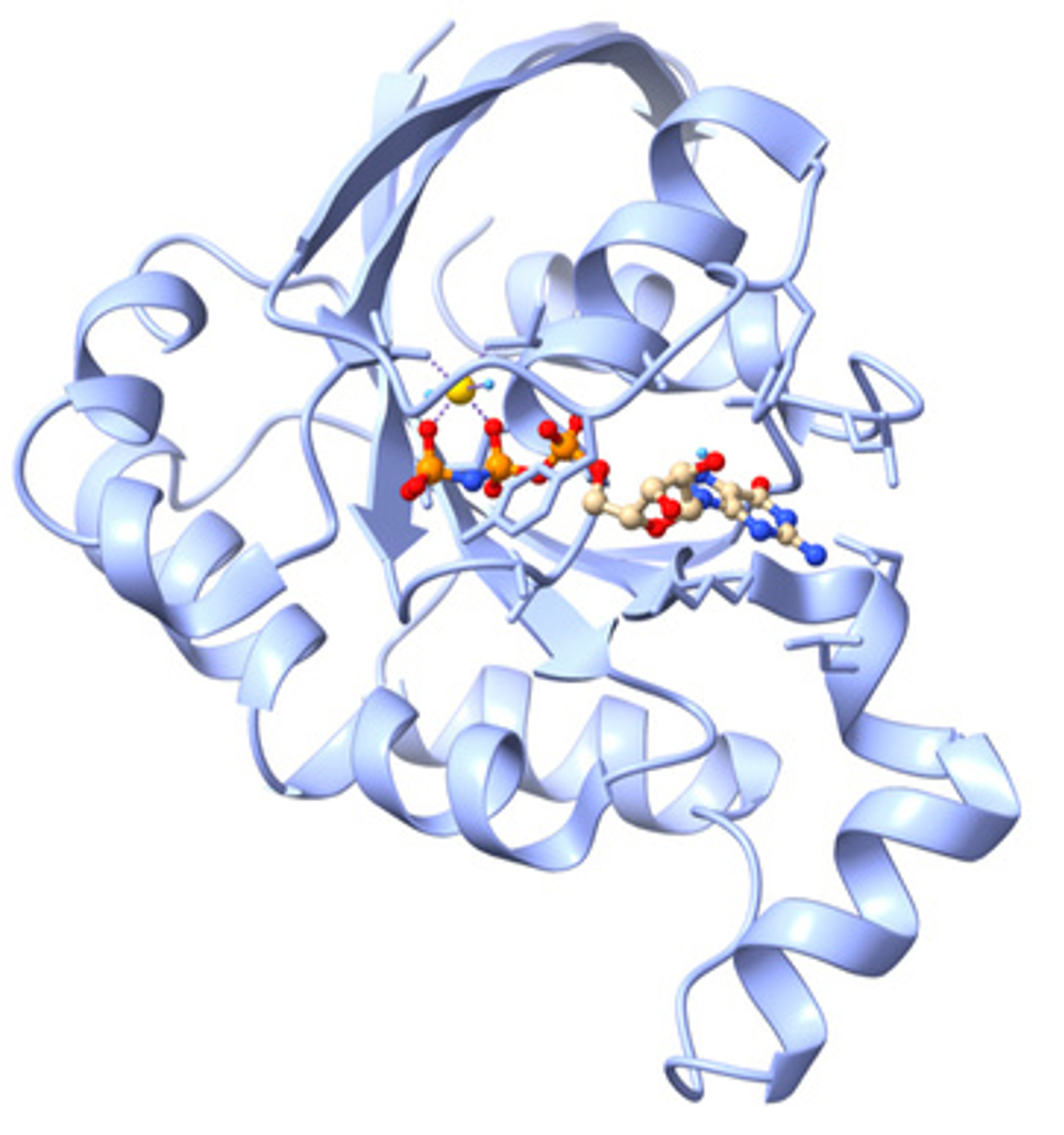
The Biophysical Chemistry group of Dr. Bayro studies the role of protein structure and dynamics in molecular mechanisms of disease to guide the design of novel therapeutic approaches. We work on understanding the inhibition of Rac1, a small GTPase whose activation cycle is associated with cancer. This nucleotide-binding protein undergoes changes in conformation and dynamics upon activation in a process that has become the target of drug design. In collaboration with cell biologists and medicinal chemists, our group aims to elucidate the mechanism of novel anti-cancer drugs by characterizing the atomic-level structure and dynamics of Rac1-inhibitor complexes. Experimental nuclear magnetic resonance (NMR) spectroscopy data is combined with molecular modeling and docking analysis to validate current hypotheses of Rac1 activation and its inhibition. Students learn to produce proteins by recombinant DNA techniques and site-directed mutagenesis, acquire protein NMR spectra in high-field magnets, extract conformation and dynamics information, and incorporate experimental data with molecular modeling for protein-drug analysis.
Dr. Jose A. Rodriguez-Martinez
DNA-binding preferences of transcription factors
Transcription factors are sequence specific DNA-binding proteins that regulate gene expression by targeting specific regions of the genome. Determining a transcription factor’s intrinsic DNA-binding preferences is a critical step for decoding gene regulatory networks that control cell function. REU fellows in the Rodriguez-Martinez lab will engage in multiple aspects of the characterization of transcriptionfactors, including the generation of large datasets of transcription factor binding preferences. REU fellows will learn molecular cloning methods, protein expression/purification, DNA-binding assays, and the use of bioinformatics tools to integrate “big data” to identify putative binding sites across genomes. All this work is guided by the Rodriguez-Martinez lab mission to uncover the molecular mechanisms by which proteins and small molecules bind to DNA and RNA to interpret genetic information.
Dr. Imilce Rodriguez Fernandez
Identification of the genes in L. plantarum capable of activating the transcription factorNrf2/CncC in the fly intestine
Dr. Rodriguez’s lab aims at understanding interactions between the gut microbiome and the intestinal epithelium. Lactobacillus species are commensal bacteria commonly found in the fly and mammalian small intestines and are generally considered probiotic agents. Studies using flies mono-associated with Lactobacillus plantarum (L. plantarum) have uncovered significant insights into host-microbial interactions. Recent findings have shed light onto how L.plantaruminteract with the gut epithelium without resulting in an activation of the immune response. It has been shown that L.plantarum can activate the transcription factor Nrf2/CncC in the host’s intestine and this could explain some of its beneficial effects. However, which genetic pathways in L. plantarum are needed for this activation are not known. IQ-Bio-REU students will work towards identifying genes in L.plantarum that are needed for the activation of the transcription factor Nrf2/CncC in the fly intestine. REU fellows will gather experience in working with the fruit fly Drosophila melanogaster, performing bacterial mutagenesis, and designing and performing genetic screenings in Drosophila and L.plantarum. Additionally, students will be able to learn fluorescence microscopy, bacterial culturing, handling and dissection of transgenic flies, and large-scale data analysis from genetic screens.
Dr. Alfredo Ghezzi
Epigenetic Mechanisms of Neuroadaptation and Addiction
Dr. Ghezzi’s research revolves around the molecular basis of neural adaptation. Through the integration of molecular genomics, behavioral analyses, and neurophysiology, his lab strives to resolve how the nervous system utilizes a finite number of genes to carefully modulate its activity and adapt to an ever-changing environment. A central component of this effort is directed to understanding the mechanisms of transcriptional memory that perpetuate these adaptations through the interplay between environmental signals, gene expression, and neural function. One fascinating example of neural adaptation is drug addiction. Drug addiction is a complex neurobiological condition characterized by compulsive and escalating drug use, and is believed to arise in part from drug-induced neural adaptations that counteract the drug effects and contribute to the uncontrolled urge to consume the drug. Students in the Ghezzi Lab will combine the powerful Drosophila genetic model and a range of genomic techniques to uncover and categorize the epigenetic regulatory mechanisms that orchestrate the response to different drugs of abuse and other environmental insults.
Dr. Jose E. Garcia-Arraras
Nervous System and Intestinal Regeneration
Our lab is interested in regenerative processes focusing on intestinal and nervous system regeneration. For this we use a novel model system; the sea cucumber Holothuria glaberrima, an echinoderm with amazing regenerative capacities. We have a large data bank of transcriptomic analyses of normal and regenerating tissues. In addition, we have completed a chromosome level genome of the species. We are now using these data to identify and characterize the molecular basis of regeneration. Experiments in the lab range from bioinformatic analyses and identification of genes and gene families to bench work where the expression of individual or clusters of genes are determined and their role in the regeneration process is determined.
Dr. Trevor Fristoe

Understanding fundamental dynamics that shape the geographic distribution of species is critical for predicting responses to global change and in disentangling the factors that generate and maintain global biodiversity patterns. The Fristoe Ecology Lab works in diverse systems, from plants through birds and mammals to humans, to reveal how species persist and coexist across diverse and dynamic environments to understand the distribution of life on Earth. Our work is macroecological in scope and bridges scales of biological organization – from physiology, traits, and behavior of individuals to spatiotemporal dynamics of populations to the distributions of species and assembly of communities across and among continents. We address questions by leveraging large ecological, environmental, geographic, and phylogenetic datasets unified across disparate sources and through the creative application of computational and statistical techniques. Within these themes, REU fellows in the Fristoe lab will choose between projects that provide opportunities to learn skills such as the management, statistical analysis, and visualization of big data in ecology, phylogenetic comparative methods, geographic analyses (GIS), or simulation approaches.
Dr. Dorta-Estremera
Multi-parameter flow cytometry in Immuno-oncology.
Dr. Dorta-Estremera is developing a research program to dissect immunological mechanisms involved in treatment resistance on human papillomavirus-induced cancers, as well as dissecting the interaction between microbial and immune cells in mucosal tissues and the tumor microenvironment to identify immune and microbial biomarkers that could be targeted to improve current therapies, especially in Hispanics. By using mouse preclinical models and/or clinical samples, students will learn to perform aseptic techniques, in vitro cultures, sample processing for flow cytometry acquisition, and flow cytometry analysis. Possible projects may include immune phenotyping of cervicovaginal samples from patients with cervical dysplasia and cervical cancer, as well as immune phenotyping of oropharyngeal mouse tumors after treatment with antibiotics and/or probiotics. Projects will generate multi-parameter immunological data to be associated with disease, ethnicity, HPV type and/or treatment response.
Dr. Kelcie Chiquillo

Unraveling epigenetic pathways to unveil invasive seagrass dynamics
In the Chiquillo Lab, we utilize ecological monitoring tools and advanced genetic techniques to explore the ecology and evolution of marine organisms across the Caribbean. One aspect of our research centers on the hypothesis that environmental changes, alongside species interactions, contribute to phenotypic plasticity and invasive traits in an invasive seagrass, Halophila stipulacea. To test this hypothesis, we conducted a reciprocal transplant experiment involving H. stipulacea and native seagrass Syringodium filiforme to assess how species interactions influence their acclimation and adaptation. Preliminary results have unveiled that the invasive seagrass demonstrated comparable growth in both reciprocal transplant sites while displaying altered vegetative traits suggestive of a distinct growth strategy. We aim to dive deeper into these findings to investigate if these changes in vegetative traits are linked to epigenetic modifications underlying the plant’s invasiveness. The IQ BIO REU participant will leverage bioinformatics tools to assemble the current Halophila genome from raw sequencing data, predict potential genes within this assembly, and create a gene transfer file (GTF) by annotating these genes with their genomic coordinates and functional elements. While the genome has been sequenced, the assembly process from raw reads remains to be conducted. Additionally, they will align methylome sequences to identify specific methylated genes associated with invasive characteristics. This project offers a unique opportunity to gain invaluable skills in genome assembly, GTF annotation, and methylome alignment, crucial for unraveling the mysteries behind invasive seagrass dynamics.
